Table of Contents
Introduction. 3
Research problem.. 8
Research aim.. 8
Research Objectives. 8
Methodology. 8
Data collection. 9
Results. 9
Differentiation. 9
Infection. 13
PUBLIC Gene expression. 16
Discussion. 18
References. 21
To investigate differentiation and infection resistance genes in macrophages
Introduction
A macrophage is a form of phagocyte which is responsible for the detection, engulfment, and destruction of pathogens and apoptotic cell types (Yang, Dai, Tang, Le, and Yao, 2017). Macrophages are formed by differentiating monocytes, which leave the blood due to tissues damage or infection, and transform into macrophages (Weavers, Evans, Martin, and Wood, 2016). Macrophages/monocytes are essential cells that engulf pathogens that cause chronic infections or viral infection in the organism. In the case of inflammation, monocytes go through serial alterations to become macrophages. Phagocytosis is the expression used to designate the engulfing and destruction of infectious or defective cells. The cellular enzymes present within the macrophage kill the ingested component. Many macrophages serve as predators, killing dead or necrotic cells whereas others engulf microbes to provide host immunity. Macrophage also acts as an Antigen Presenting Cells (APCs). An APC is a kind of an immune cell that detects, engulfs and indicates an infection to the adaptive immune system. These APCs can phagocytose the pathogen, once it is recognized, and break it down in antigen fragments. Antigen fragments are then transferred to the APC surface, where they act as markers for other immune cells (Pei, Liu, Zhou, and Fan, 2020).
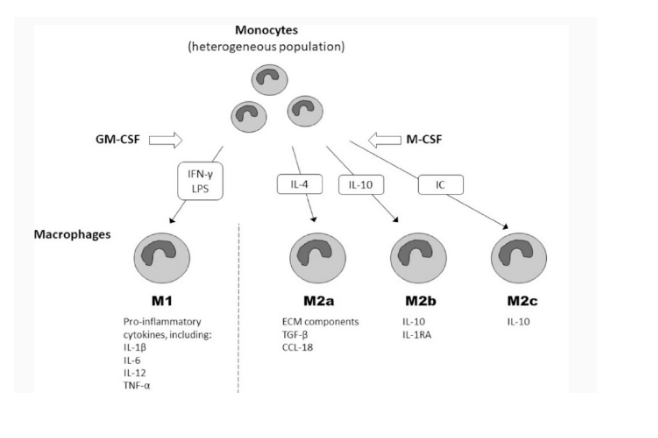
Monocytes were considered the primary role of sensing the environment and replenishing the stream of tissue macrophages (Barrila et al., 2017). The macrophages are classified as non-activated (M0), pro-inflammatory (M1), or anti-inflammatory (M2) subsets which play a distinct role in inflammation initiation and differentiation (Figure 14). Such variability also occurs at the level of the monocytes as they may also lead to pro- inflammatory as well as anti-inflammatory phenotypes. Growth factors, like GM-CSF and M-CSF, possess a critical place in their activation: GM-CSF induces the differentiation of “pro-inflammatory” monocytes to M1 macrophages, whereas M-CSF controls the differentiation of the “anti-inflammatory” subset of monocytes to M0 macrophages with functional and phenotypic properties similar to M2 (Salerno-Goncalves et al., 2019). Monocyte-to-macrophage differentiation requires global transcriptome shifts, which are closely regulated by diverse transcriptional regulators as well as signaling pathways in humans. In nutshell, macrophages are completely differentiated cells that, in reaction to dynamically changing cell environment, show considerable phenotypic plasticity though. Recognized subsets of monocytes exhibit separate pathophysiological roles. Classic inflammatory monocytes are empowered with an array of Toll-like receptors (TLRs) and scavenger receptors which recognize pathogen-associated molecular patterns (PAMPs) and remove microorganisms, fats, and apoptotic cells (Arslan et al, 2017).
(Figure 1: Activation of macrophages)
(Source: Orekhov et al., 2019)
Macrophages are important for the ability of the inherent immune response to destroy and kill intruding microbes during infection, as they merge powerful anti-microbial activities with the capacity to cause local inflammation as well as a systemic immune reaction. Phagocytosis and intracellular phagocyte destruction of bacteria is a key mechanism for the defense of the innate immune response throughout infectivity. Microbes such as bacteria are primarily destroyed by phagocytosis followed by intracellular digestion, and it is now been identified that this core anti-bacterial function may also result in phagocyte apoptosis. While apoptosis of macrophage has been reported more frequently during the onset of virulent microorganisms that have adjusted to intracellular life, such as Salmonella, Mycobacteria much research has also indicated macrophage elimination following phagocytosis of extracellular bacteria like Escherichia coli or Streptococci which are immediately eliminated upon onset (Behar, nd Briken, 2019). Many pathogens have also developed the ability of these specialized cells to cause programmed cell death. Several times it is documented that contact with pyogenic, extracellular bacterial contributes to apoptosis phagocyte death (Mendoza-Coronel, and Castanon-Arreola, 2016). In addition to enhancing inflammation and inducing the immune system, macrophages also perform a significant anti-inflammatory function and by releasing cytokines, they can reduce immune reactions.
Cytokine release in host cells is strongly linked to the bacterial survival and infectivity. Macrophages are critical in the process of pathogenesis of Salmonella infections. Most bacterial pathogens which have coevolved with its hosts also show a remarkable level of adaptation, allowing them to modulate and sometimes manipulate a variety of cellular host processes. Mechanisms for the death of salmonella-induced macrophage cells were the topic of uncertainty. Whereas some groups recorded rapid death of the macrophage following salmonella infection, others recorded the need for sustained infection (24 hours) for cell death. Cell-type expression of gene through macrophage differentiation and activity is the outcome of complicated transcriptional regulation by interplaying endogenous transcription variables with exterior signals, as well as by networking specific transcription factors. When salmonellae invade macrophage cells, they produce cytotoxins within these host cells (Yeung et al, 2019). The numerous phagocytic receptors present on the macrophage plasma layer identify Salmonella-expressed structures and compounds and the phagocytic processes activated by such receptors utilize specific procedures and signal transduction routes. Contrary to many other pathogens, however, Salmonella can survive inside of macrophages by adjusting to this specific intracellular environmental niche. Salmonella expresses virulence factors to cope with this hostile condition in order to survive in the host and to prevent clearance by the host immune system. In Salmonella, the virulence genes are accountable for pathogenicity and infection are predominantly located in the distinctive 40 kb chromosomal region known as Salmonella pathogenicity islands (SPI), which were acquired by horizontal transfer of gene. SPI-1 and SPI-2 are recognized as pathogenicity islands that encode their own Type III (T3SS) secretion processes to transmit bacterial effector proteins to the organism (Mendoza-Coronel, and Castanon-Arreola, 2016). The T3SS encoded for SPI-1 is extracellularly active. The SPI-1 regulates intrusion in the non-phagocytic cells, which is essential for immune response to the intestines. The SPI-2 pathogenic genes are presented intracellularly and are important for bacterial survival in systemic infections as well as macrophages. The SPI-1 secretion mechanism regulates bacterial intrusion into host cells (epithelial) whereas Salmonella duplication within host cells and also the systemic infections includes the SPI-2. The mutant strains of SPI-2 are significantly attenuated, showing a 104-fold reduction in LD50 (50% lethality dose) virulence in the murine salmonellosis prototype and disrupted intracellular replication as well as macrophage survival. In addition, the SPI-2 region comprises huge chunks of horizontally acquired genes that have a greater A+T presence than other sections of Salmonella DNA. Histone-like nucleoid formulating protein (H-NS) binds the AT-rich DNA to silence the binding codes (Maikho et al, 2018). Silencing facilitated by H-NS may shield Salmonella from the negative effect of over-expression of SPI-2 virulence genes at incorrect times (Wang et al, 2017).
Expression of particular transcription variable is likely to be modulated transcriptionally during differentiation process. Multiple transcription factors like EGRs, NFkB, IRFs which regulate the production or function of different stages of macrophages (Jeon et al, 2020). U937 cells are commonly seen as a model to examine a number of monocyte and macrophage based biological processes (Ares et al, 2019). The cell line U937 can be caused by treatment with either or both phorbol myristate acetate (PMA) or 1,25-dihydroxyvitamin D3 (VD) to differentiate along the macrophage pathway (Marcuello et al, 2018). Following differentiation, U937 cells, through the expression of various genes, acquire a wide repertoire of macrophage activity. With LPS (E. coli lipopolysaccharide), differentiated U937 cells may also be stimulated to imitate the immune response of the macrophages (Chen et al, 2017). The key macrophages that convey this transcript are also fitted with an improved defense mechanisms. Also essential is the downregulation of immune responses by anti-inflammatory cytokines through macrophage deactivation (Aluwi et al, 2017). The principal cause of TNF-α and IL-10 (anti inflammatory cytokines) is activated monocytes and macrophages. Fuggetta et al, (2019) that HLA-B27 may possess a place in directly regulating microbe-host cell exchanges during infection by salmonella, additional to earlier known functions of MHC class I molecules during the antigen presentation.
The study intends to understand and examine the developments of differentiation and infection resistant genes in macrophages. This study examines the how vitamin D3 (1, 25-dihydroxyvitamin D3), or PMA (phorbol 12-myristate 13-acetate) can triggerU937 monocytes differentiation into a phenotype similar to macrophage. In the lab, a PhD student Josephine Duncombe-Moore, has carried out genetic screens in U937. The first asked the question whether there are certain genes which when mutated, altered the ability of U937 cells to differentiate into macrophages. The second screen took the mature macrophages and asked if there were any genes which, when mutated, would protect macrophages from Salmonella infection– or make them more susceptible. These screens produced two lists of mutant genes relating to differentiation and infection. The study intends to examine and explore the mutated genes that affect U937 differentiation into macrophage-like cells. In my project, I have taken these lists and queried publically available gene expression datasets to verify the importance of these genes in these processes. The study aims to compare the available differentiation gene data with the values of original gene expression which are retrieved from publicly available GEO database. As genes are crucial for differentiation and the gene expression changes during differentiation, therefore, the results of this study are based on the comparison of the differentiation and infection gene data. The study aims to present the potential genes that can be harnessed in the situation of infection and be of great value in treating and eliminating infections and can function as an alternative to killing pathogens with antibiotics (infection resistance genes).
Research problem
Research aim
To investigate differentiation and infection resistance genes in mutant macrophage cells (U937 cells)
Research Objectives
1. To examine the genes which altered the ability of U937 cells to differentiate into macrophages due to mutation
2. To examine the genes in the mature macrophages which are susceptible to Salmonella infection
3. To examine the publically available gene expression datasets to verify the importance of genes involved in differentiation and infection
Methodology
The study employs data analysis technique in which the data is gathered from secondary sources that are the previously published literature. Comparisons between various experimental conditions with distinct gene expression profiles (for instance, different time points with activation and differentiation) have the ability to recognize co-regulated and/or interconnected genes.
Gene data set 1: It is the gene detection in the process of differentiation in the STM U937 (Salmonella Typhimurium) cells. The data set presents the gene expression values when the cells are differentiated into macrophages. The data set contains the values of mutated genes of the U937 cells. ABCA1 (ATP binding cassette subfamily A member 1), TNFSF15 (TNF superfamily member 15), PARP9 (Poly (ADP-ribose) polymerase family member 9), TMEM51-AS1 (TMEM51 antisense RNA 1), SPP1 (Secreted phosphoprotein 1), SKIL (SKI like proto-oncogene), and MTO1 (Mitochondrial tRNA translation optimization 1).
The study employs data analysis technique in which the data is gathered from secondary sources that are the previously published literature. Comparisons between various experimental conditions with distinct gene expression profiles (for instance, different time points with activation and differentiation) have the ability to recognize co-regulated and/or interconnected genes.
Gene data set 1: It is the gene detection in the process of differentiation in the STM U937 (Salmonella Typhimurium) cells. The data set presents the gene expression values when the cells are differentiated into macrophages. The data set contains the values of mutated genes of the U937 cells. ABCA1 (ATP binding cassette subfamily A member 1), TNFSF15 (TNF superfamily member 15), PARP9 (Poly (ADP-ribose) polymerase family member 9), TMEM51-AS1 (TMEM51 antisense RNA 1), SPP1 (Secreted phosphoprotein 1), SKIL (SKI like proto-oncogene), and MTO1 (Mitochondrial tRNA translation optimization 1).
Gene data set 2: It is the gene detection in the process of infection. The data set containing the values of genes (C5orf60, LGALS12, SLC7A11, SERPINB2, ZNF770, TRIM60P14 Gene(Pseudogene), SQLE, MANSC1) during infection. The infection gene value is measured both in the original and control environments.
By comparing the gene sets 1 with 2 can present the mutations in genes which prevent the differentiation or facilitate it. Additionally, by comparing the gene sets 2 with transcriptional data can illustrate which mutations in genes are acting protective or facilitating susceptibility. These categories represent their functional associations, and may help define common mechanisms of transcriptional control. Transcription factors can be accountable for controlling the genes in the same subset within these groups of genes. Hence, the data will be analyzed with the help of spreadsheets and presented in the forms of tables and charts.
Data collection
The data for this research is collected with the help Arrayexpress and NCBI GEO online sources. GEO is a public data repository for functional genomics which supports MIAME-compliant data submission. It accepts array- and sequence-centered data. Tools are offered to assist users in querying and uploading studies and curated profiles on gene expression. GEO Data Sets and Profiles form a part of the Entrez database network of NCBI. Like with all other repositories, data of interest will easily be found by entering search terms boxes of the GEO Data Sets or GEO Profiles.
The infection of macrophages by different species of bacteria, virus, fungi etc is retrieved from GEO database. With the help of GEO search term: ‘macrophage infection’, ‘macrophage fungal infection’, the gene code samples were extracted for Staphylococcus aureus, TLR agonist R848, Mycobacterium tuberculosis, LPS, African swine fever virus, Dengue fever, Toxoplasma, Cryptococcus, Staphylococcus aureus (NCBI GEO, 2020). The gene variations (retrieved from GEO database) are recorded in the sub sets of ABCA1 (ATP binding cassette subfamily A member 1), TNFSF15 (TNF superfamily member 15), PARP9 (Poly (ADP-ribose) polymerase family member 9), TMEM51-AS1 (TMEM51 antisense RNA 1), SPP1 (Secreted phosphoprotein 1), SKIL (SKI like proto-oncogene), and MTO1 (Mitochondrial tRNA translation optimization 1).
Results
Differentiation
The Gene data (set 1) contains the experimental gene data where the gene values are recorded in the Original as well as control sets. The specific genes provided in data set 1 which indicated mutation are – ABCA1, TNFSF15, PARP9, TMEM51-AS1, SPP1, SKIL, and MTO1. The gene data represents the gene expression values when the cells are differentiated into macrophages. On the onset of differentiation, the U937 cells acquire a wide range of macrophage activity with the concerted expression of various genes. The results from the gene data set 1 for the specific genes are displayed as Table 1. These are the values of gene expression which are mutated to facilitate differentiation and convert the undifferentiated U937 cells into differentiated macrophages.
| ABCA1 | TNFSF15 | PARP9 | TMEM51 | SPP1 | MTO1 | SKIL | |
| 6.78315 | 6.484485 | 7.2204 | 7.33536 | 5.765364 | 6.446134 | 2664.63 | |
| 10.349084 | 10.29613 | 10.37787 | 9.67265 | 14.7058 | 6.257344 | 393.0117 | |
| 3.565934 | 3.811645 | 3.157467 | 2.33729 | 8.940436 | -0.18879 | -2271.62 | |
| 0.0001 | 1.5 | 3 | 336 | 3 | 74342.5 | 21 | (Undifferentiation) |
| 1259.666667 | 1397 | 1571.667 | 8652.5 | 29650.83 | 19057.33 | 2059.667 | (Differentiation) |
| 7.93861E-08 | 0.001074 | 0.001909 | 0.038833 | 0.000101 | 3.900992 | 0.010196 | (X- axis) |
(Table 1: Gene expressions in the genetic screens (original vs control) in U937 by Josephine Duncombe-Moore)
(Source: Provided experimental data)
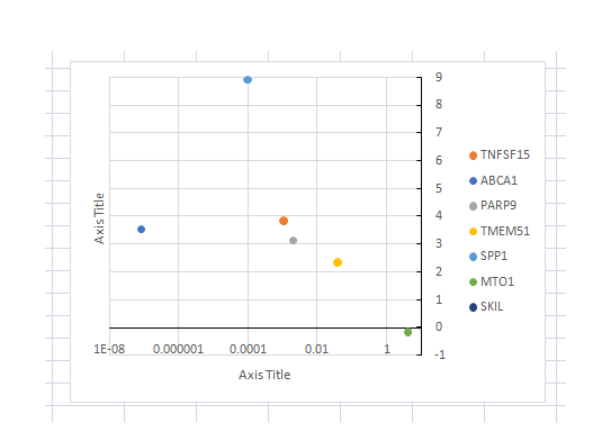
(Figure 2: Graph of Gene expressions in the genetic screens (original vs control) in U937 by Josephine Duncombe-Moore)
(Source: Provided experimental data)
The amount of gene expression of the mutated genes which derived the differentiation in the U937 cells is displayed as Figure 3. It can be clearly observed that the value of the gene expression has been increased in the process of differentiation to convert the cells into macrophages.
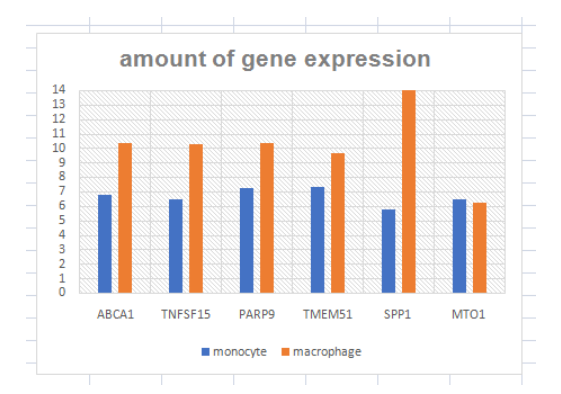
(Figure 3: Amount of Gene expressions in undifferentiated cells)
(Source: Learner (derived from experimental data))
The figure 4 presents the graph for cells which were differentiated but not infected with the original data which presented the undifferentiated moncoytes. It is a clear indication of how many cells undergo differentiated on treatment with Vit D or PMA. The differentiated cells are capable to differentiate themselves into anti-inflammatory subsets or inflammatory subsets. U937 cells are considered to vary by PMA along a lineage of monocytes / macrophages and this line of cells provides a good model for cell differentiation (López, & Urcuqui-Inchima, 2018). Differentiation results in changes in cells which make them more functional. The key changes or improvement is the phagocytic ability to activate the monocytes. The available gene data shows that catalase posses a crucial place in the process of differentiation of U937 cells toward a macrophage/monocyte lineage. The gene expression of certain genes which is derived from the gene data set 1 is displayed as Figure 8. It can be seen that the except for MTO1, there is significant changes in the gene expressions for ABCA1, TNSFS15, APAp9, TMEM51, and significant changes in the SPP1. Several genes in the experimental data do not change their value of expression in the process of macrophage differentiation.
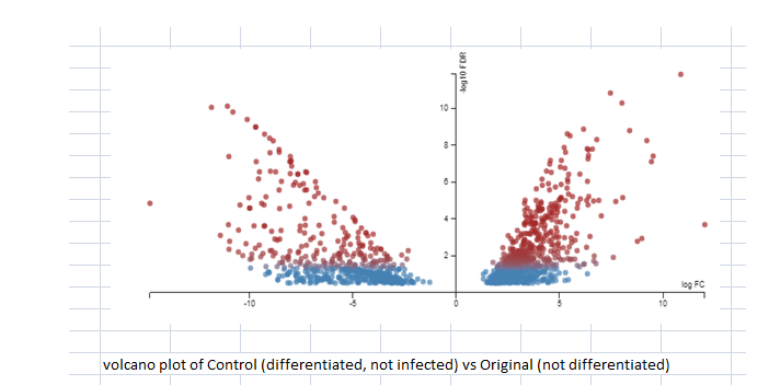
(Figure 4: Comparison between differentiated and undifferentiated; FDR threshold set at 0.3; No fold change threshold set)
(Source: Experimental data)
Genetic expression of differentiated or activated cells as opposed to undifferentiated U937 cells was determined. U937 cells included are known to be similar to functional monocytes as demonstrated by the lower amount of genes expressed differently upon VitD3 differentiation, while becoming an appropriate model approach for investigating gene regulatory systems in macrophage differences (Billings et al, 2016). Array assessment of differentiated PMA and LPS activated cells showed a number of macrophage-intrinsic genes, innate immunity and inflammation. Differing expression of gene are deemed noteworthy whenever the sample vs control normalized strength ratio was or < -2 (down) or > 1.9 (up), respectively (. At PMA segregation, 320 genes were regulated upwards and 179 genes regulated down. Also, 337 genes were regulated further up while 126 genes were regulated down. Additionally, a sum of 595 genes was regulated upwards and 324 regulated down. 275 additional genes were up regulated after 2 hours of subsequent LPS stimulation while 105 additional genes were regulated down. For VitD3 variation, only 9 genes were regulated significantly up, whereas 12 genes were regulated downwards. Out of them are some genes considered to be linked to the cycle of differentiation, and also macrophage and monocyte functions. This includes proteins deemed to be active in the process of differentiation (CDK inhibitor p21), cytokine transporters such as A2 M, chemokines such as CCL3 and IP10, and matrix metalloproteinase proteins like MMP9 and MMP7 active in structural as well as adhesive matrix proteolysis (Dallenga et al, 2017). Nonetheless, enhanced expression of chemokines and cytokines is one of the phenotypic attibutes of differentiated macrophages.
The data comparison of the gene expression in cells which are differentiated but not infected with the original cells which are undifferentiated, an understanding of the monocyte maturation into macrophages is established. It is notable that variations in gene expression rates between differentiation cells and macrophages frequently resulted from a particular down-regulation of gene expression in one type of cell over another. For instance, the expression of C5orf60, was down-regulated in differentiation cells and macrophages relative to monocytes, but its expression remained higher in macrophages than in differentiation cells, and its macrophage expression remained comparable to that seen in monocytes, resulting in a macrophage-specific expression pattern of these genes.
Infection
The Gene data (set 2) presents the gene which protect against or make susceptible to Salmonella infection of C5orf60, LGALS12, SLC7A11, SERPINB2, ZNF770, TRIM60P14 Gene (Pseudogene), SQLE, MANSC1) during infection. The infection gene value is measured both in the original and control environments. The genes that changed expression are considered as the monocytes which matured into macrophages (Tian, Xie, Ding, and Zhou, 2018). It is the gene detection in the process of differentiation in the STM U937 (Salmonella Typhimurium) cells. After comparing the gene data set with transcriptional data, it is observed that the mutated genes LGALS12, SLC7A11, SERPINB2, ZNF770, SQLE, MANSC1 (Table 2) derived infection. Hence, it can be said that with STM (Salmonella Typhimurium), the differentiated U937 cells are stimulated to imitate the inflammatory response of the activated macrophages.
| LGALS12 | SLC7A11 | SERPINB2 | ZNF770 | SQLE | MANSC1 |
| 9.764116 | 5673.502 | 120.30728 | 918.6936 | 1124.9534 | 121.98122 |
| 18.89654 | 16359.6 | 5854.926 | 907.8556 | 978.5736 | 61.64046 |
| 1.935305 | 2.88351004 | 48.6664315 | 0.9882028 | 0.8698792 | 0.505327459 |
| 11 | 0.5 | 2.83333333 | 311 | 24 | 23 |
| 0.5 | 6.83333333 | 83 | 1.3333333 | 1.5 | 0.001 |
| 0.045455 | 13.6666667 | 29.2941176 | 0.0042872 | 0.0625 | 4.34783E-05 |
(Table 2: Amount of Gene expressions in undifferentiated cells)
(Source: Learner (derived from experimental data))
By comparing the original and control data for specific gene expression, differentiation and the macrophage maturation process can be understood. For instance, the infection is seen in two instances which imply that at four instances the differentiated macrophage is able to control the infection and thus removed the infection in 4 instances (Figure 5). Similarly, SLC7A11 expression has presented that the differentiated cells are infected in all instances when compared the control and original data. Nonetheless, MANSC1 and ENSG00000264918 gene expressions have clearly demonstrated that the differentiated cells are able to contain the infection at all instances (Figure 6 and 7) whereas, SERPINB2 gene expression presents that the differentiated cells are unable to contain the infection in the given instances in original as well as control.
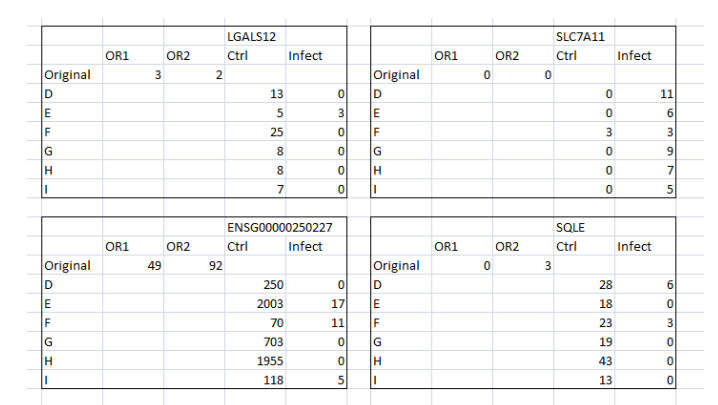
(Figure 5: Values of mutated Gene expressions (Original and control) in the context of infection)
(Source: Infection data)
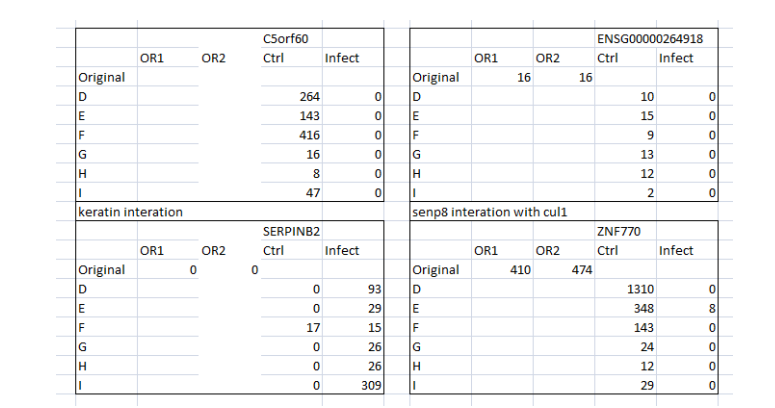
(Figure 6: Values of mutated Gene expressions (Original and control) in the context of infection)
(Source: Infection data)
The level of differentiation in U937 cells with STM is compared with the original gene data values which are derived from the GEO data sets to investigate which genes are capable of controlling the infection in the cells and which are not. With this comparison, the genes can be identified and recorded for their application against the infection by salmonella. Following infection, the host genes involved in the biosynthesis, degradation, and intracellular lipid transport were significantly regulated. Upon infection, genes involved in the cholesterol biosynthesis process increased in expression such as SQLE.
The key findings of the comparison of the differentiation data and the infection data it is noted that:
SQLE increased in Control compared to Original
MANSC1 increased in Control compared to Original
SQLE (squalene epoxidase) – It catalyzes the first oxygenation stage in sterol biosynthesis and is one of the rate-restricting enzymes in this bio-pathway. SQLE increased in the control as compared to the original which represents the maturation of monocytes into macrophage which is uninfected (or have the ability to control the infection).
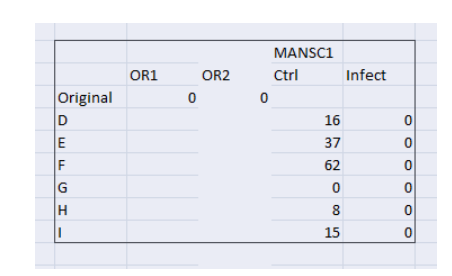
(Figure 7: Values of mutated Gene expressions (Original and control) in the context of infection)
(Source: Infection data)
PUBLIC Gene expression
The collected data (from GEO) for the mutated genes in the differentiation process is displayed below as Figure in the fold change value in the infected and uninfected macrophages.
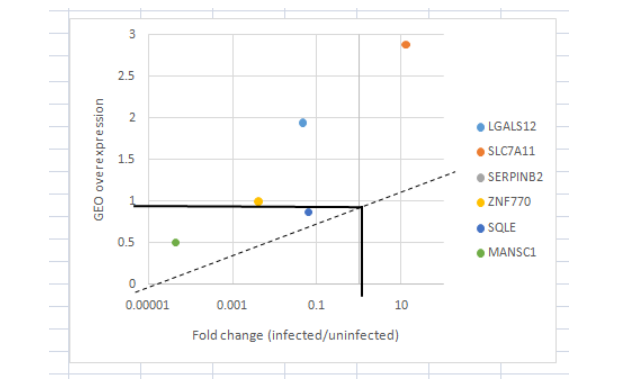
(Figure 8: Amount of Fold change in Gene expressions)
(Source: Learner (derived from GEO dataset))
The GEO data base has been accessed top compare the findgs and find out the genes which are responsible for driving infection in different microorganisms (table 3).
| Hcar2 | cryptococcus | Aqp9 | Toxoolasma | MMP12 | LPS | DUSP5 | LPS | |
| Hcar2 | staphylococcus | AQP9 | LPS | Nfkbia | Toxoolasma | Edn1 | Toxoolasma | |
| Ptges | cryptococcus | Arl5b | Toxoolasma | NFKBIA | LPS | EDN1 | LPS | |
| Ptges | staphylococcus | ARL5B | LPS | Nr4a3 | Toxoolasma | Egr1 | Toxoolasma | |
| Slc2a1 | cryptococcus | Ccl5 | Toxoolasma | NR4A3 | LPS | EGR1 | LPS | |
| Slc2a1 | staphylococcus | CCL5 | LPS | Ptger4 | Toxoolasma | Egr3 | Toxoolasma | |
| Hcar2 | cryptococcus | Ccr7 | Toxoolasma | PTGER4 | LPS | EGR3 | LPS | |
| Hcar2 | staphylococcus | CCR7 | LPS | Ptgs2 | Toxoolasma | Etv3 | Toxoolasma | |
| Hcar2 | Toxoplasma | Ccrl2 | Toxoolasma | PTGS2 | LPS | ETV3 | LPS | |
| Bhlhe40 | cryptococcus | CCRL2 | LPS | Rel | Toxoolasma | Gfi1 | Toxoolasma | |
| Bhlhe40 | Toxoplasma | Cd274 | Toxoolasma | REL | LPS | GFI1 | LPS | |
| Clec7a | cryptococcus | CD274 | LPS | Socs3 | Toxoolasma | Id2 | Toxoolasma | |
| Clec7a | Toxoplasma | Cd40 | Toxoolasma | SOCS3 | LPS | ID2 | LPS | |
| Cox7a1 | cryptococcus | CD40 | LPS | Tnfrsf9 | Toxoolasma | Il1rn | Toxoolasma | |
| Cp | Toxoplasma | Cd83 | Toxoolasma | TNFRSF9 | LPS | DUSP2 | LPS | |
| Hcar2 | cryptococcus | CD83 | LPS | Traf1 | Toxoolasma | Dusp5 | Toxoolasma | |
| Hcar2 | Toxoplasma | Cxcl2 | Toxoolasma | TRAF1 | LPS | |||
| IL1RN | LPS | CXCL2 | LPS | INHBA | LPS | |||
| Inhba | Toxoolasma | Dusp2 | Toxoolasma | Mmp12 | Toxoolasma |
(Table 3: Macrophage Genes responsible for infection in different species of microorganisms)
(Source: Derived from GEO dataset)
Hence, it can be asserted that the findings are not in alignment with the publically accessed data for the mutated genes in macrophages which can facilitate infection. It is due to the fact that the specific genes which drive infection in the macrophages in the experimental data is different from the genes which are responsible for infection in other specifies. In this study, the heterogeneity of monocyte-macrophage (differentiation), and gene expressions that control differentiation at transcription level is addressed. The gene expression in differentiated cells and Macrophages were examined separately. The findings further suggest that process of differentiation of macrophages can lead to the acquisition of varying signaling capabilities in the two types of cells. Genes associated with apoptosis and IL6 signaling were upregulated in both types of cells. Also, the receptor signaling pathway is up‐regulated especially in the macrophages. Numerous genes encoding recognized macrophage and DC surface marker molecules, such as CD14 and CD163 for macrophages and CD1a, DCSIGN, CD1e, and the DC lysosome-linked membrane protein (CD208) for differentiated cells, have been found to be expressed within various differentiation stages (Aurass et al., 2018). These differentially expressed genes are primarily involved in the Salmonella infectivity, DNA synthesis, energy transfer and oxidation, and nutrient material intake and metabolism, etc. Amongst these upregulated genes were those located on the SPI-2, Salmonella pathogenicity, plays a critical role in the intracellular survival and replication of Salmonella. The results indicate that particular virulence factors in the pathogens can decrease the intracellular growth in the host cells.
Discussion
With the findings of this study it can be asserted that a considerable increase in phagocytic activity was seen in cells which matured into macrophages. Throughout the differentiation progression and subsequent activation, transcriptional regulators are recognized to be implicated in monocytic differentiation (such as EGR1, or MAFB) are continuously up regulated. Earlier unknown variables (such as BRI, TCF7L2 family, etc) have also been shown to be controlled considerably and constantly up.FOS, STAT3, IRF family, and SMAD display substantial up regulation upon LPS stimulation, thus confirming their classical involvement in inflammatory response macrophage activityne of the NFkB family ‘s key physiological functions is the formation and operating of the immune response by controlling the synthesis of cytokines and also antimicrobial effectors and genes that modulate cellular survival, differentiation, and also proliferation (Yang et al, 2017). cREL, NFkB1 and NFkB2 are consistently induced significantly due to LPS stimulation, consequently verifying the relevance of the U937 model system. Similar populations of macrophages from various body tissues display functional and morphological phenotypes (López, and Urcuqui-Inchima, 2018). M-CSF separated macrophages are well known to be very similar to peritoneal macrophages. M-CSF modified macrophages were produced from mainly monocytes as a corresponding aspect of PMA-differentiated U937 cells, but were subsequently activated with LPS. In addition, unlike PMA-differentiated U937 cells when up regulation was seen, monocyte-derived macrophages present down regulation of TNFalpha (Zhao et al, 2018).
The results indicated the mutated genes such as SQLE, MANSC1 are responsible for facilitating the Salmonella infection in the differentiated U937 cells. The other genes found from the GEOO data base which can drive infection in other microorganizams are:
Cryptococcus: Hcar2, Ptges, Slc2a1, Bhlhe40, Clec7a
Staphylococcus: Ptges, Egr3, Il1rn, Traf1, Ccl5, Aqp9, Slc2a1, Hcar2
Toxoolasma: Aqp9, Arl5b, Ccl5, Ccr7, Ccrl2, Cd274, Cd40, Cd83, Cxcl2, Dusp2, Dusp5, Edn1, Egr1, Egr3, Etv3
LPS: AQP9, ARL5B, CCL5, CCR7, CCRL2, CD274, CD40, CD83, CXCL2, DUSP2, DUSP5, EDN1, EGR1, EGR3, ETV3, GFI1, ID2, IL1RN, INHBA, MMP12, NFKBIA, NR4A3, PTGER4, PTGS2, REL, SOCS3, TNFRSF9, TRAF1
The identified genes possess a crucial role in the protein formation and immunity response which regulates the infection in the microorganism. For instance, the SLC2A1 gene gives instructions for manufacturing a protein known as the glucose transporter protein type 1 (GLUT1). CLEC7A (C-type lectin domain family 7, member A) encodes a member of the C-type lectin/C-type lectin-like domain (CTL/CTLD) superfamily. CD83 encodes protein which is a single-pass type I membrane protein and member of the immunoglobulin superfamily of receptors. The encoded protein is involved in the regulation of antigen presentation. Edn1 (Endothelin 1) – encodes a protein which is proteolytically processed to release a secreted peptide termed endothelin 1 (Flannagan, Kuiack, McGavin, and Heinrichs, 2018).
Hence, in order to understand the cellular process for the capacity of the macrophage to resist the infection, it can be emphasized that the functional and phenotypic macrophage plasticity is important for effective tissue injury healing and the removal of infection. However, the entire process is onset with the presence of pathogenic environment and autophagy in the macrophages. Peng et al., 2018 demonstrated that autophagy induction is important for the differentiation of macrophages/monocytes. Both pharmacological and siRNA approaches induced monocytes inhibits the phenomenon of autophagy due to which the monocytes failed to differentiate into functional macrophages (Elitas, M., & Sengul, 2020). M1 macrophages are involved in combating and destroying pathogens or cancer cells at the inflammation site, and then further removing dead cells and cell waste. M1 macrophages are active in attracting monocytes and lymphocytes to the injury site (Zhou et al., 2020). In reaction to anti-inflammatory stimuli, which are able to resolve inflammation and promoting tissue repair and remodeling in a pathogen-free microenvironment, M1 macrophages do not vanish when the injured site is clear, but undergo a phenotypic transition to M2 macrophages. M1-M2 conversion also helps keep the inflamed site from producing an unnecessary influx of pro-inflammatory immune cells. Activation with GM-CSF in monocytes upregulates the antigen-presenting role, phagocytosis, anti-microbial activity, and development of pro-inflammatory cytokines (IL-1β, IL-6 , IL-8, TNF-α), as well as growth factors (M-CSF, GM-CSF). A global transcriptome study of the macrophages caused by GM-CSF revealed up-regulation of 340 genes primarily responsible for lipid metabolism, antigen presentation, as well as innate immune signaling comprising of macrophage-specific surface receptors/markers like CD14, CD163, C5R1, CSF3R, GDF15, and FcγR1A. (Haque et al., 2020). Several studies have established the phenomenon of differentiation of monocytes cells into macrophages due to the presence of pathogenic environment. Authors (Qi, et al, 2017) suggested that Macrophages possess critical roles in the regulation of inherent immune reactions caused by viruses and are believed to be engaged in pathogenesis of bacterial infections. In addition, the rates of IL-10 and TGF-Î23 mRNA in both subsets of superinfection were marginally upregulated relative to the community of single LPS stimulation or virus only infection. Similarly, Curto, Simões, Riley, & Martinez (2016) indicated that a phenotypical distinction amongst a non-pathogenic as well as a pathogenic SFG group occurs in their respective capacity to intrude in macrophage-like cells.
To conclude it can be asserted that Macrophage phagocytosis of apoptotic cells may have anti-inflammatory effects which can help to overcome inflammation and minimize tissue damage. Autophagy by macrophage is crucial to host responses towards bacterial infections, transmitting intracellular pathogens to lysosomes for degradation while regulating inflammation to minimize damage to the host. The findings of the study presented the various mutated genes in macrophages which are responsible for the infection in the different microorganisms such as Staphylococcus. Comprehending the differentiation mechanism of the monocytes will probably offer a potential therapeutic benchmark for inflammatory monocytosis and can aid in developing effective antibiotics.
References
Aluwi, M.F.F.M., Rullah, K., Yamin, B.M., Leong, S.W., Bahari, M.N.A., Lim, S.J., Faudzi, S.M.M., Jalil, J., Abas, F., Fauzi, N.M. and Ismail, N.H.(2016). Synthesis of unsymmetrical monocarbonyl curcumin analogues with potent inhibition on prostaglandin E2 production in LPS-induced murine and human macrophages cell lines. Bioorganic & medicinal chemistry letters, 26(10), pp.2531-2538.
Ares, M.A., Sansabas, A., Siqueiros-Cendón, T., Rascón-Cruz, Q., Rodríguez-Valverde, D., Jarillo-Quijada, M., Alcántar-Curiel, M.D., Rosales-Reyes, R., Torres, J. and De la Cruz, M.A. (2019). The interaction of Klebsiella pneumoniae with lipid rafts-associated cholesterol increases macrophage-mediated phagocytosis due to down regulation of the capsule polysaccharide. Frontiers in cellular and infection microbiology, 9, p.255.
Arslan, B. A., Isik, F. B., Gur, H., Ozen, F., & Catal, T. (2017). Apoptotic effect of Nigella sativa on human lymphoma U937 cells. Pharmacognosy Magazine, 13(Suppl 3), S628.
Aurass, P., Düvel, J., Karste, S., Nübel, U., Rabsch, W., & Flieger, A. (2018). glnA truncation in Salmonella enterica results in a small colony variant phenotype, attenuated host cell entry, and reduced expression of flagellin and SPI-1-associated effector genes. Applied and Environmental Microbiology, 84(2).
Barrila, J., Yang, J., Crabbé, A., Sarker, S. F., Liu, Y., Ott, C. M., … & Davis, R. R. (2017). Three-dimensional organotypic co-culture model of intestinal epithelial cells and macrophages to study Salmonella enterica colonization patterns. Npj Microgravity, 3(1), 1-12.
Behar, S.M. and Briken, V. (2019). Apoptosis inhibition by intracellular bacteria and its consequence on host immunity. Current Opinion in Immunology, 60, pp.103-110.
Billings, E.A., Lee, C.S., Owen, K.A., D’Souza, R.S., Ravichandran, K.S. and Casanova, J.E. (2016). The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Science signaling, 9(413), pp.ra14-ra14.
Chen, C.L., Chien, S.C., Leu, T.H., Hans, I., Harn, C., Tang, M.J. and Hor, L.I. (2017). Vibrio vulnificus MARTX cytotoxin causes inactivation of phagocytosis-related signaling molecules in macrophages. Journal of biomedical science, 24(1), p.58.
Curto, P., Simões, I., Riley, S. P., & Martinez, J. J. (2016). Differences in intracellular fate of two spotted fever group Rickettsia in macrophage-like cells. Frontiers in cellular and infection microbiology, 6, 80.
Dallenga, T., Repnik, U., Corleis, B., Eich, J., Reimer, R., Griffiths, G.W. and Schaible, U.E. (2017). M. tuberculosis-induced necrosis of infected neutrophils promotes bacterial growth following phagocytosis by macrophages. Cell host & microbe, 22(4), pp.519-530.
Elitas, M., & Sengul, E. (2020). Quantifying Heterogeneity According to Deformation of the U937 Monocytes and U937-Differentiated Macrophages Using 3D Carbon Dielectrophoresis in Microfluidics. Micromachines, 11(6), 576.
Flannagan, R.S., Kuiack, R.C., McGavin, M.J. and Heinrichs, D.E. (2018). Staphylococcus aureus uses the GraXRS regulatory system to sense and adapt to the acidified phagolysosome in macrophages. MBio, 9(4).
Fuggetta, M.P., Zonfrillo, M., Villivà, C., Bonmassar, E. and Ravagnan, G. (2019). Inflammatory microenvironment and adipogenic differentiation in obesity: the inhibitory effect of theobromine in a model of human obesity in vitro. Mediators of inflammation, 2019.
Haque, S., Kodidela, S., Sinha, N., Kumar, P., Cory, T. J., & Kumar, S. (2020). Differential packaging of inflammatory cytokines/chemokines and oxidative stress modulators in U937 and U1 macrophages-derived extracellular vesicles upon exposure to tobacco constituents. Plos one, 15(5), e0233054.
Jeon, E.H., Park, T.S., Jang, Y., Hwang, E., Kim, S.J., Song, K.D., Weinstein, D.A., Lee, Y.M., Park, B.C. and Jun, H.S. (2020). Glucose-6-phosphate transporter mediates macrophage proliferation and functions by regulating glycolysis and mitochondrial respiration. Biochemical and Biophysical Research Communications.
López, J. V., & Urcuqui-Inchima, S. (2018). Synergism between phorbol-12-myristate-13-acetate and vitamin D3 in the differentiation of U937 cells to monocytes and macrophages. Morphologie, 102(338), 205-218.
Maikho, T., Patwardhan, R.S., Das, T.N., Sharma, D. and Sandur, S.K. (2018). Cold atmospheric plasma-modulated phorbol 12-myristate 13-acetate-induced differentiation of U937 cells to macrophage-like cells. Free radical research, 52(2), pp.212-222.
Marcuello, M., Mayol, X., Felipe-Fumero, E., Costa, J., López-Hierro, L., Salvans, S., Alonso, S., Pascual, M., Grande, L. and Pera, M. (2018). Modulation of the colon cancer cell phenotype by pro-inflammatory macrophages: A preclinical model of surgery-associated inflammation and tumor recurrence. Plos one, 13(2), p.e0192958.
Mendoza-Coronel, E. and Castanon-Arreola, M. (2016). Comparative evaluation of in vitro human macrophage models for mycobacterial infection study. Pathogens and Disease, 74(6).
NCBI GEO. (2020). Comparative gene expression in response to various inflammatory stimuli in vitro: infection-mediated versus systemic inflammation. Retrieved from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126525
NCBI GEO. (2020). Expression data from WT and Klf3 KO BMDMs treated with LPS. Retrieved from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121646
Orekhov, A. N., Orekhova, V. A., Nikiforov, N. G., Myasoedova, V. A., Grechko, A. V., Romanenko, E. B., … & Chistiakov, D. A. (2019). Monocyte differentiation and macrophage polarization. Vessel Plus, 3(10).
Pei, X., Liu, M., Zhou, H. and Fan, H. (2020). Screening for phagocytosis resistance-related genes via a transposon mutant library of Streptococcus suis serotype 2. Virulence, 11(1), pp.825-838.
Peng, M., Tabashsum, Z., Patel, P., Bernhardt, C., & Biswas, D. (2018). Linoleic acids overproducing Lactobacillus casei limits growth, survival, and virulence of Salmonella Typhimurium and enterohaemorrhagic Escherichia coli. Frontiers in microbiology, 9, 2663.
Qi, X., Liu, C., Li, R., Zhang, H., Xu, X., & Wang, J. (2017). Modulation of the innate immune-related genes expression in H9N2 avian influenza virus-infected chicken macrophage-like cells (HD11) in response to Escherichia coli LPS stimulation. Research in veterinary science, 111, 36-42.
Salerno-Goncalves, R., Kayastha, D., Fasano, A., Levine, M. M., & Sztein, M. B. (2019). Crosstalk between leukocytes triggers differential immune responses against Salmonella enterica serovars Typhi and Paratyphi. PLoS neglected tropical diseases, 13(8), e0007650.
Tian, X., Xie, G., Ding, F. and Zhou, X. (2018). LPS-induced MMP-9 expression is mediated through the MAPKs-AP-1 dependent mechanism in BEAS-2B and U937 cells. Experimental lung research, 44(4-5), pp.217-225.
Wang, H.C., Chen, C.W., Yang, C.L., Tsai, I.M., Hou, Y.C., Chen, C.J. and Shan, Y.S. (2017). Tumor-associated macrophages promote epigenetic silencing of gelsolin through DNA methyltransferase 1 in gastric cancer cells. Cancer immunology research, 5(10), pp.885-897.
Weavers, H., Evans, I.R., Martin, P. and Wood, W. (2016). Corpse engulfment generates a molecular memory that primes the macrophage inflammatory response. Cell, 165(7), pp.1658-1671.
Yang, L., Dai, F., Tang, L., Le, Y. and Yao, W. (2017). Macrophage differentiation induced by PMA is mediated by activation of RhoA/ROCK signaling. The Journal of toxicological sciences, 42(6), pp.763-771.
Yeung, A.T., Choi, Y.H., Lee, A.H., Hale, C., Ponstingl, H., Pickard, D., Goulding, D., Thomas, M., Gill, E., Kim, J.K. and Bradley, A. (2019). A Genome-Wide Knockout Screen in Human Macrophages Identified Host Factors Modulating Salmonella Infection. Mbio, 10(5), pp.e02169-19.
Zhao, Y.L., Lu, Z.Y., Zhang, X., Liu, W.W., Yao, G.D., Liu, X.L., Liu, W., Wu, Q.J., Hayashi, T., Yamato, M. and Fujisaki, H. (2018). Gelatin promotes cell aggregation and pro-inflammatory cytokine production in PMA-stimulated U937 cells by augmenting endocytosis-autophagy pathway. The International Journal of Biochemistry & Cell Biology, 95, pp.132-142.
Zhou, A., Li, J., Xu, Z., Ni, J., Guo, J., Yao, Y. F., & Wu, W. (2020). Whole-Genome Comparative and Pathogenicity Analysis of Salmonella enterica subsp. enterica serovar Rissen. G3: Genes, Genomes, Genetics.




